Metabolism plays a role in virtually all cellular processes. Understanding how nutrients, metabolites, and byproducts flow through intracellular pathways (i.e., metabolic flux) is invaluable for elucidating disease mechanisms and designing new therapies. To quantitatively visualize pathway activity we employ stable isotope-labeled metabolic tracers and detect isotope enrichment in metabolites using chromatography coupled to mass spectrometry. The amount of isotope-labeled atoms that are incorporated into biomass or other pathways tells us about how our tracer was metabolized. Finally, to account for the interconnected nature of metabolic networks we apply systems-level analyses of these data, since flux through one reaction is affected by many other pathways.
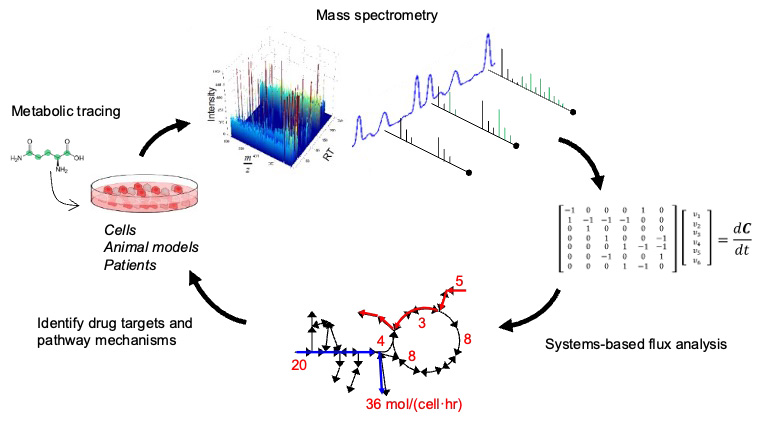
The general approach used in our lab involves the 1) application of stable isotope tracers to living systems; 2) analysis of how these tracer molecules are metabolized across tissues, cells, and organelles; and 3) modeling to better interpret results and conclusions in order to better understand disease mechanisms and identify strategies for intervention.
Cancer metabolism
Oncogenes, tumor suppressors, and the tissue microenvironment induce profound changes in cancer cell behavior and biochemistry. Tumor cells rewire metabolic pathways to grow, survive, and metastasize to other organs. These biochemical alterations can drive new therapeutic vulnerabilities. By exploring how cancer cell genetics and metabolism are intertwined, we hope to identify key nutritional and/or metabolic dependencies of cancer cells and develop new strategies for slowing tumor progression in patients. For example, we have examined how hypoxia regulates pathways fueling lipid synthesis in cancer cells, the redox sensitivity of cancer cells harboring oncogenic isocitrate dehydrogenase 1 (IDH1) mutations, and the role of the liver kinase B1 (LKB1) tumor suppressor in controlling metabolism in response to stress. We have also explored how serine, glycine, and mitochondrial amino acid metabolism support cancer progression through their impact on circulating sphingolipids and membrane lipid diversity in tumor cells.
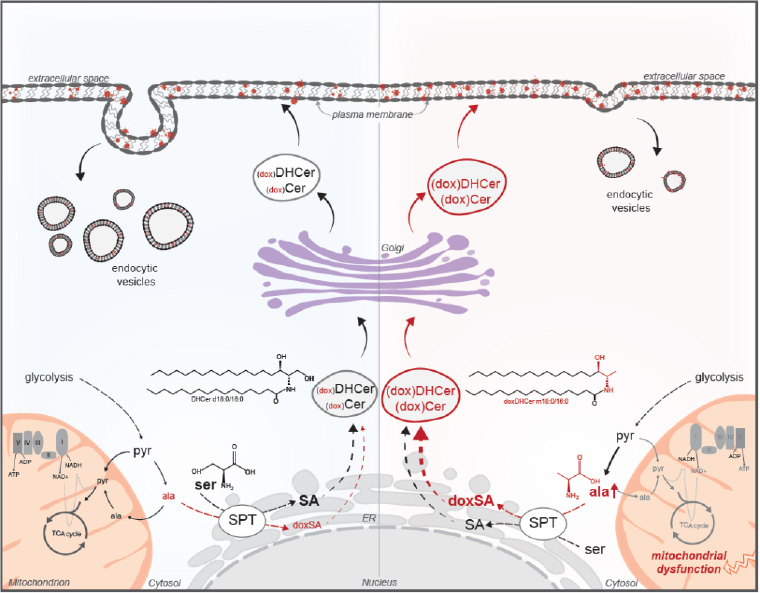
Depiction of how mitochondrial dysfunction influences amino acid balancing and sphingolipid diversity in cancer cells growing under anchorage-independent conditions. The accumulation of alanine or depletion of serine results in synthesis of 1-deoxysphingolipids, which reduce plasma membrane-associated vesicle trafficking.
Current efforts aim to elucidate how fluxes are coordinated across organelles throughout the lipidome to support cancer growth and survival. Numerous coordinated and redundant pathways are required to maintain pools of complex membrane lipids important for cell signaling and other processes. We are tracing and modeling how these biochemical pathways are regulated in distinct cell populations within tumors.
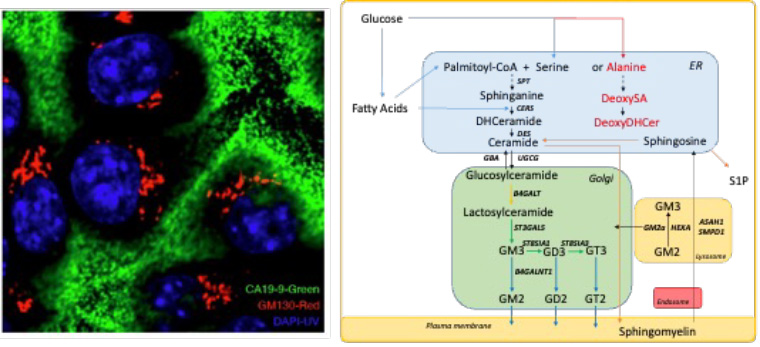
Left: Pancreatic ductal adenocarcinoma cells synthesize and secrete glycans (green) within the golgi apparatus (red). Right: Complex lipid biosynthesis proceeds through the endoplasmic reticulum (ER), golgi, and plasma membrane, while membrane lipids and components are recycled and degraded through endolysosomal transport.
References
- Cordes et al., 1-deoxysphingolipid synthesis compromises anchorage-independent growth and plasma membrane endocytosis in cancer cells. (2022) Journal of Lipid Research
- Muthusamy et al., Serine restriction alters sphingolipid diversity to constrain tumour growth. (2020) Nature
- Badur et al., Reverse engineering the cancer metabolic network using flux analysis to understand drivers of human disease. (2018) Metabolic Engineering
- Parker and Metallo, Chasing One-Carbon Units to Understand the Role of Serine in Epigenetics. (2016) Molecular Cell
- Vacanti et al., Regulation of substrate utilization by the mitochondrial pyruvate carrier. (2014) Molecular Cell
- Metallo, Expanding the reach of cancer metabolomics. (2012) Cancer Prevention Research
- Grassian et al., Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. (2011) Genes & Development
Metabolic syndrome, obesity, and co-morbidities
Diabetes, metabolic syndrome, and obesity represent a spectrum of diseases and co-morbidities that involve dysregulation of metabolically active cells in the liver, kidney, adipose tissue, muscle, and nervous system. The breakdown of nutrient signaling and physiology between these tissues drives diabetes. While the role of carbohydrates and fat in these processes is fairly well appreciated, we and other have highlighted key mechanisms through which amino acid metabolism becomes dysregulated in diabetes and obesity. Branched-chain amino acid (BCAA) catabolism is active in diverse cell types, regulated by tissue hypoxia, and altered in diabetes. Flux in adipose tissue yields monomethyl branched-chain fatty acids (BCFAs), which may act as biomarkers of disease states. We have also discovered that insulin signaling controls serine catabolism during feeding, contributing to reduced circulating serine and glycine in diabetic animals and driving peripheral neuropathy. By applying metabolic flux analysis and other approaches to cells, animals, and humans, we hope to discover how changes in carbohydrate, protein, amino acid, and lipid metabolism contribute to metabolic syndrome and related ailments.
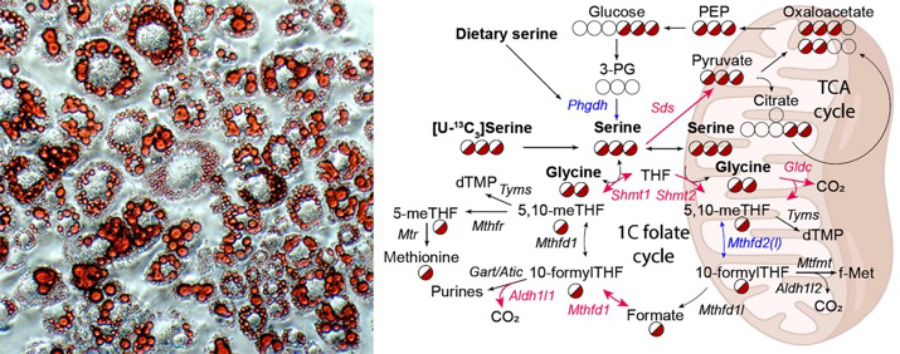
Left: Oil red O staining of cultured adipocytes highlighting lipid droplets within cells. Right: Metabolic pathways involving serine, glycine, and one carbon metabolism are dysregulated within the diabetic liver.
References
- Handzlik et al, Insulin-regulated serine and lipid metabolism drive peripheral neuropathy (2023, in press), Nature
- Wallace et al, Enzyme promiscuity drives branched-chain fatty acid synthesis in adipose tissues. (2018) Nature Chemical Biology
- Green et al, Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. (2016) Nature Chemical Biology
Neurodegenerative disorders and macular disease
The nervous system is comprised of neurons and glia which coordinate bioenergetic, catabolic, and biosynthetic pathways to meet their significant metabolic demands. Coordinated regulation of metabolic pathways using small molecules can sustain nervous system function in response to stress. Genetic defects in that occur in mitochondria or other pathways can have diverse consequences in cells and tissues. Collaborations with the Lowy Medical Research Institute focus on understanding macular telangiectasia, a rare familial disease that is characterized by altered circulating amino acids and sphingolipids.
References
- Lim et al, Progressive alterations in amino acid and lipid metabolism correlate with peripheral neuropathy in mice. (2021) Science Advances
- Kumar et al, NaCT/SLC13A5 facilitates citrate import and metabolism under nutrient-limited conditions. (2021) Cell Reports
- Cordes et al., Itaconate modulates tricarboxylic acid and redox metabolism to mitigate reperfusion injury. (2020) Molecular Metabolism
- Gantner, et al., Serine and Lipid Metabolism in Macular Disease and Peripheral Neuropathy. (2019) New England Journal of Medicine
Developing targeted approaches to study metabolism and physiology
Given our broad interest in metabolism, we are continually striving to identify or develop new methods for deciphering how metabolic pathways are wired in living systems. These include tools to study critical pathways involved in redox homeostasis, cellular bioenergetics, and lipid metabolism within distinct organelles or cell types. Application of these methods to pluripotent stem cells and their derivatives enables characterization of diverse cell lineages and disease states.
References
- Zhao et al., Combinatorial CRISPR-Cas9 Metabolic Screens Reveal Critical Redox Control Points Dependent on the KEAP1-NRF2 Regulatory Axis. (2018) Molecular Cell
- Badur et al., Enzymatic passaging of human embryonic stem cells alters central carbon metabolism and glycan abundance. (2015) Biotechnol Journal
- Lewis et al., Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. (2014) Molecular Cell